| Length 3' UTR: | 1,352 nt |
| A+T content in 3' UTR: | 0.64 |
| RNAplfold output: | [ opening energies | probabilities of being unpaired ] (whole transcript) |
| Download/Linkout: | [ download as annotated Genbank file | Linkout to  ] ] |
| ATTTA: | 0.82 fold-enrichment |
| Site 2837-2841: | ATTTA (ATTTA)
Opening energy for the core AUUUA pentamer: 0.96 kcal/mol (short range) / 1.55 kcal/mol (mid range)
Probability of being unpaired for the core AUUUA pentamer: 0.21 (short range) / 0.08 (mid range)
[ Highlight | show accessibility plot | show sequence logo | show alignment ] |
Short range interaction (W = 80, L = 40)Mid range interaction (W = 240, L = 160)Note: Accessibility values (energy needed to open secondary structures, probability of being unpaired) are calculated using RNAplfold. RNAplfold has been called with various setting for u=n, where n is the number of consecutive bases. For example, the value at position i with u=5 is the probability that i and the four preceeding nucleotides are unpaired. Taking the logarithm of this probability we can easliy derive the opening energy for this stretch of nucleoitdes. | Genomic alignmnent block extracted from 30-way MAF alignments (UCSC genome browser). | Alignment of transcripts based on the Ensembl orthology resources. | 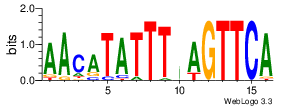 | 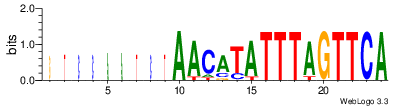 |
|
| Site 3573-3577: | ATTTA (ATTTA)
Opening energy for the core AUUUA pentamer: 1.34 kcal/mol (short range) / 2.13 kcal/mol (mid range)
Probability of being unpaired for the core AUUUA pentamer: 0.11 (short range) / 0.03 (mid range)
[ Highlight | show accessibility plot | show sequence logo | show alignment ] |
Short range interaction (W = 80, L = 40)Mid range interaction (W = 240, L = 160)Note: Accessibility values (energy needed to open secondary structures, probability of being unpaired) are calculated using RNAplfold. RNAplfold has been called with various setting for u=n, where n is the number of consecutive bases. For example, the value at position i with u=5 is the probability that i and the four preceeding nucleotides are unpaired. Taking the logarithm of this probability we can easliy derive the opening energy for this stretch of nucleoitdes. | Genomic alignmnent block extracted from 30-way MAF alignments (UCSC genome browser). | Alignment of transcripts based on the Ensembl orthology resources. | 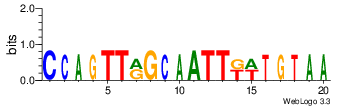 | 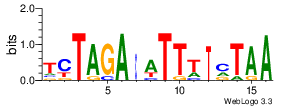 |
|
| Site 3582-3586: | ATTTA (ATTTA)
Opening energy for the core AUUUA pentamer: 0.70 kcal/mol (short range) / 0.67 kcal/mol (mid range)
Probability of being unpaired for the core AUUUA pentamer: 0.32 (short range) / 0.34 (mid range)
[ Highlight | show accessibility plot | show sequence logo | show alignment ] |
Short range interaction (W = 80, L = 40)Mid range interaction (W = 240, L = 160)Note: Accessibility values (energy needed to open secondary structures, probability of being unpaired) are calculated using RNAplfold. RNAplfold has been called with various setting for u=n, where n is the number of consecutive bases. For example, the value at position i with u=5 is the probability that i and the four preceeding nucleotides are unpaired. Taking the logarithm of this probability we can easliy derive the opening energy for this stretch of nucleoitdes. | Genomic alignmnent block extracted from 30-way MAF alignments (UCSC genome browser). | Alignment of transcripts based on the Ensembl orthology resources. | 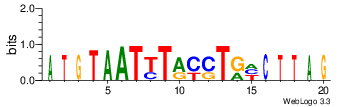 |  |
|
| Site 3801-3805: | ATTTA (ATTTA)
Opening energy for the core AUUUA pentamer: 0.57 kcal/mol (short range) / 0.91 kcal/mol (mid range)
Probability of being unpaired for the core AUUUA pentamer: 0.40 (short range) / 0.23 (mid range)
[ Highlight | show accessibility plot | show sequence logo | show alignment ] |
Short range interaction (W = 80, L = 40)Mid range interaction (W = 240, L = 160)Note: Accessibility values (energy needed to open secondary structures, probability of being unpaired) are calculated using RNAplfold. RNAplfold has been called with various setting for u=n, where n is the number of consecutive bases. For example, the value at position i with u=5 is the probability that i and the four preceeding nucleotides are unpaired. Taking the logarithm of this probability we can easliy derive the opening energy for this stretch of nucleoitdes. | Genomic alignmnent block extracted from 30-way MAF alignments (UCSC genome browser). | Alignment of transcripts based on the Ensembl orthology resources. | 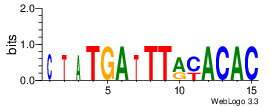 | 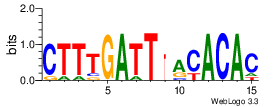 |
|
 ]
]






