

| [Home|New job|Help] |
Results
The optimal secondary structure upon hybridization is given below. It ranges from positions 24 to 27 in the longer sequence and from positions
4 to 7 in the short sequence.
Total free energy of binding: -2.66 kcal/mol
Energy from duplex formation: -3.00 kcal/mol
Opening energy for the longer sequence: 0.13 kcal/mol
Opening energy for the shorter sequence: 0.21 kcal/mol
You can access the RNAup output here.
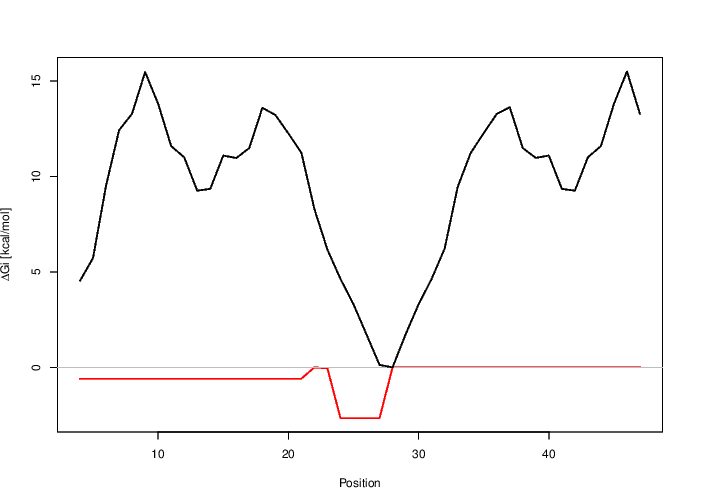
Graphical output
The plot below shows the interaction free energy (RED)  Gi and the energy needed to open
existing structures in the longer sequence (BLACK) for the target region (top) and the whole range (bottom). You can download the top plot as [EPS|PDF|IMAGE CONVERTER].
You can download the bottom plot as [EPS|PDF|IMAGE CONVERTER].
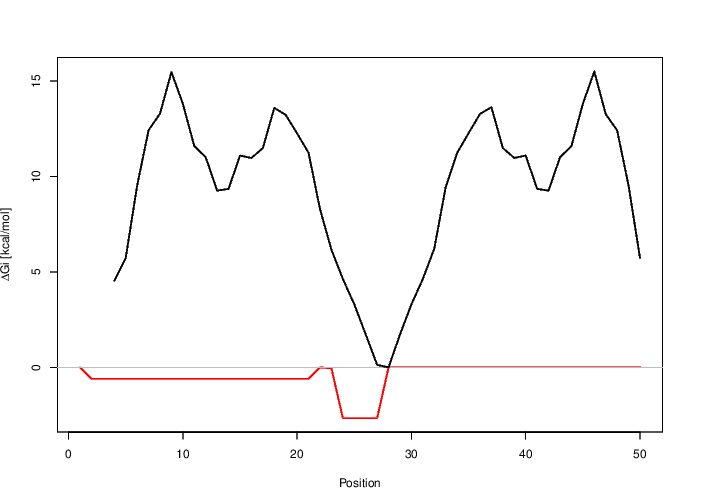
Gi and the energy needed to open
existing structures in the longer sequence (BLACK) for the target region (top) and the whole range (bottom). You can download the top plot as [EPS|PDF|IMAGE CONVERTER].
You can download the bottom plot as [EPS|PDF|IMAGE CONVERTER].
Results have been computed using RNAup 2.6.3. An equivalent command line call would have been
RNAup -b -d2 --noLP -c 'S' < sequences.fa > RNAup.out
RNA parameters are described in
Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A 101(19):7287-92.
If you find these results helpful for your work you may want to cite:

|
Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL.
The Vienna RNA Websuite. Nucleic Acids Res. 2008 |
U.Mueckstein, H. Tafer, J. Hackermueller, S.H. Bernhart, P.F. Stadler, and I.L. Hofacker "Thermodynamics of RNA-RNA Binding." Bioinformatics, 22(10), pp 1177-1182, 2006